Introduction
While analyzing with SCTK, there might be excessive data that should be removed. Here we briefly talk about the way to clean up the unwanted information.
To view detailed instructions on how to delete data, please select ‘Interactive Analysis’ for deletion through the shiny application or ‘Console Analysis’ for deletion through the R console from the tabs below:
Workflow Guide
SCTK provides a page that allows users to delete existing data, including count matrices (assays), cell (column) annotation, feature (row) annotation, dimension reduction and subsets. To delete data, open up the “Delete Single Cell Data” tab as shown above.
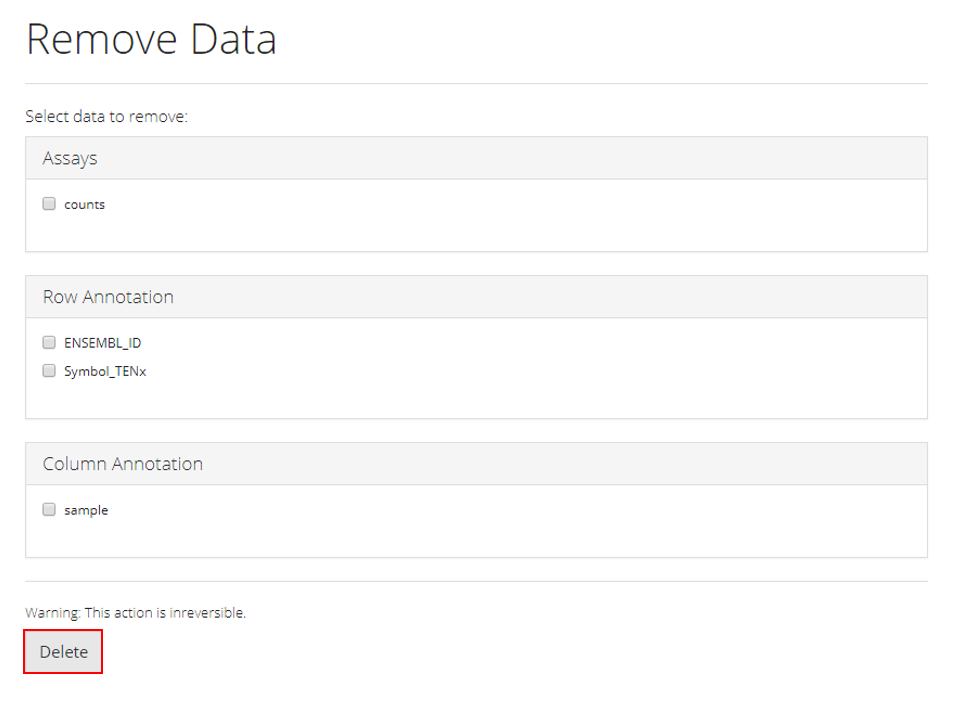
The operation here is intuitive. As shown in the screenshot above, users can easily delete any unwanted or useless information by checking the corresponding checkboxes. Users have to be careful when checking the data because this deletion operation is not reversible. Finally, click on “Delete” button to finish the operation.
SCTK uses the SingleCellExperiment object as the container of users’ datasets.
For example, some data has been computed and stored in the object as below:
# Get a SingleCellExperiment object
library(singleCellTK)
sce <- importExampleData("pbmc3k")
sce <- scaterlogNormCounts(sce, "logcounts")
sce <- subsetSCERows(sce, index = seq(100), returnAsAltExp = TRUE, altExpName = "subset")
sce <- scaterPCA(sce, reducedDimName = "PCA")This data can be removed as below by simply setting the required slots to NULL:
## Remove a certain cell annotation (colData)
colData(sce)$Tissue_status <- NULL
## Remove a certain feature annotation (rowData)
rowData(sce)$Symbol <- NULL
## Remove a certain feature expression matrix (expData)
expData(sce, "logcounts") <- NULL
## Remove a certain feature subset (altExp)
altExp(sce, "subset") <- NULL
## Remove a certain dimension reduction (reducedDim)
reducedDim(sce, "PCA") <- NULL