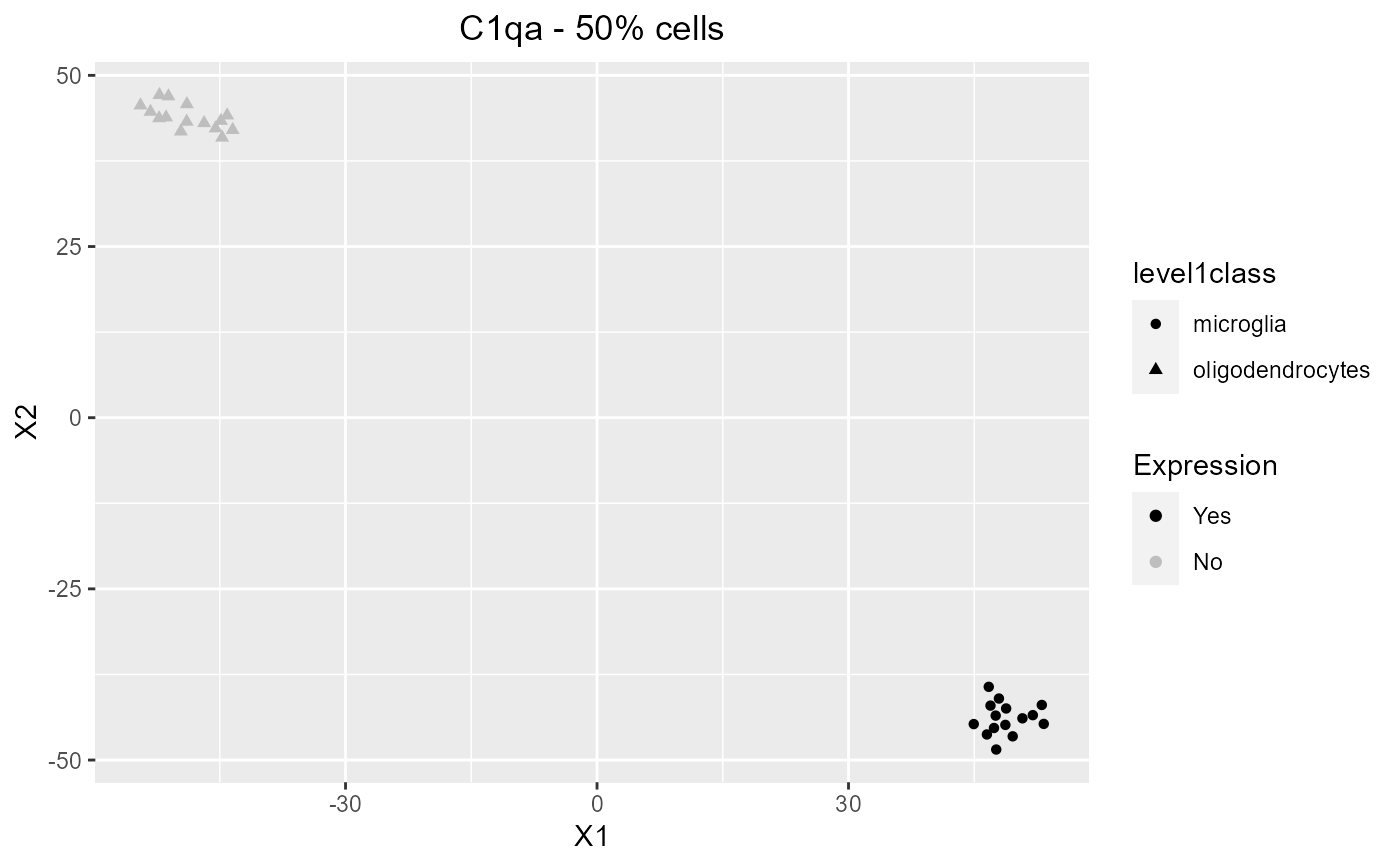
Given a set of genes, return a ggplot of expression values.
Source:R/plotBiomarker.R
plotBiomarker.RdGiven a set of genes, return a ggplot of expression values.
plotBiomarker( inSCE, gene, binary = "Binary", shape = "No Shape", useAssay = "counts", reducedDimName = "PCA", x = NULL, y = NULL, comp1 = NULL, comp2 = NULL )
Arguments
| inSCE | Input SingleCellExperiment object. |
|---|---|
| gene | genelist to run the method on. |
| binary | binary/continuous color for the expression. |
| shape | shape parameter for the ggplot. |
| useAssay | Indicate which assay to use. The default is "logcounts". |
| reducedDimName | results name of dimension reduction coordinates obtained from this method. This is stored in the SingleCellExperiment object in the reducedDims slot. Required. |
| x | PCA component to be used for plotting(if applicable). Default is first PCA component for PCA data and NULL otherwise. |
| y | PCA component to be used for plotting(if applicable). Default is second PCA component for PCA data and NULL otherwise. |
| comp1 | label for x-axis |
| comp2 | label for y-axis |
Value
A Biomarker plot
Examples
data("mouseBrainSubsetSCE") plotBiomarker(mouseBrainSubsetSCE, gene="C1qa", shape="level1class", reducedDimName="TSNE_counts")