Compare a result object to COSMIC V3 Signatures; Select exome or genome for SBS and only genome for DBS or Indel classes
Source:R/compare_results.R
compare_cosmic_v3.RdCompare a result object to COSMIC V3 Signatures; Select exome or genome for SBS and only genome for DBS or Indel classes
Usage
compare_cosmic_v3(
musica,
model_id,
sample_type,
modality = "SBS96",
result_name = "result",
metric = "cosine",
threshold = 0.9,
result_rename = deparse(substitute(model_id)),
decimals = 2,
same_scale = FALSE
)Arguments
- musica
A
musicaobject.- model_id
The name of the model containing the signatures to compare.
- sample_type
exome (SBS only) or genome
- modality
Compare to SBS, DBS, or Indel. Default
"SBS96"- result_name
Name of the result list entry. Default
"result".- metric
One of
"cosine"for cosine similarity or"jsd"for 1 minus the Jensen-Shannon Divergence. Default"cosine".- threshold
threshold for similarity
- result_rename
title for plot user result signatures
- decimals
Specifies rounding for similarity metric displayed. Default
2.- same_scale
If
TRUE, the scale of the probability for each signature will be the same. IfFALSE, then the scale of the y-axis will be adjusted for each signature. DefaultTRUE.
Examples
data(res)
compare_cosmic_v3(res,
model_id = "res", modality = "SBS96",
sample_type = "genome", threshold = 0.8
)
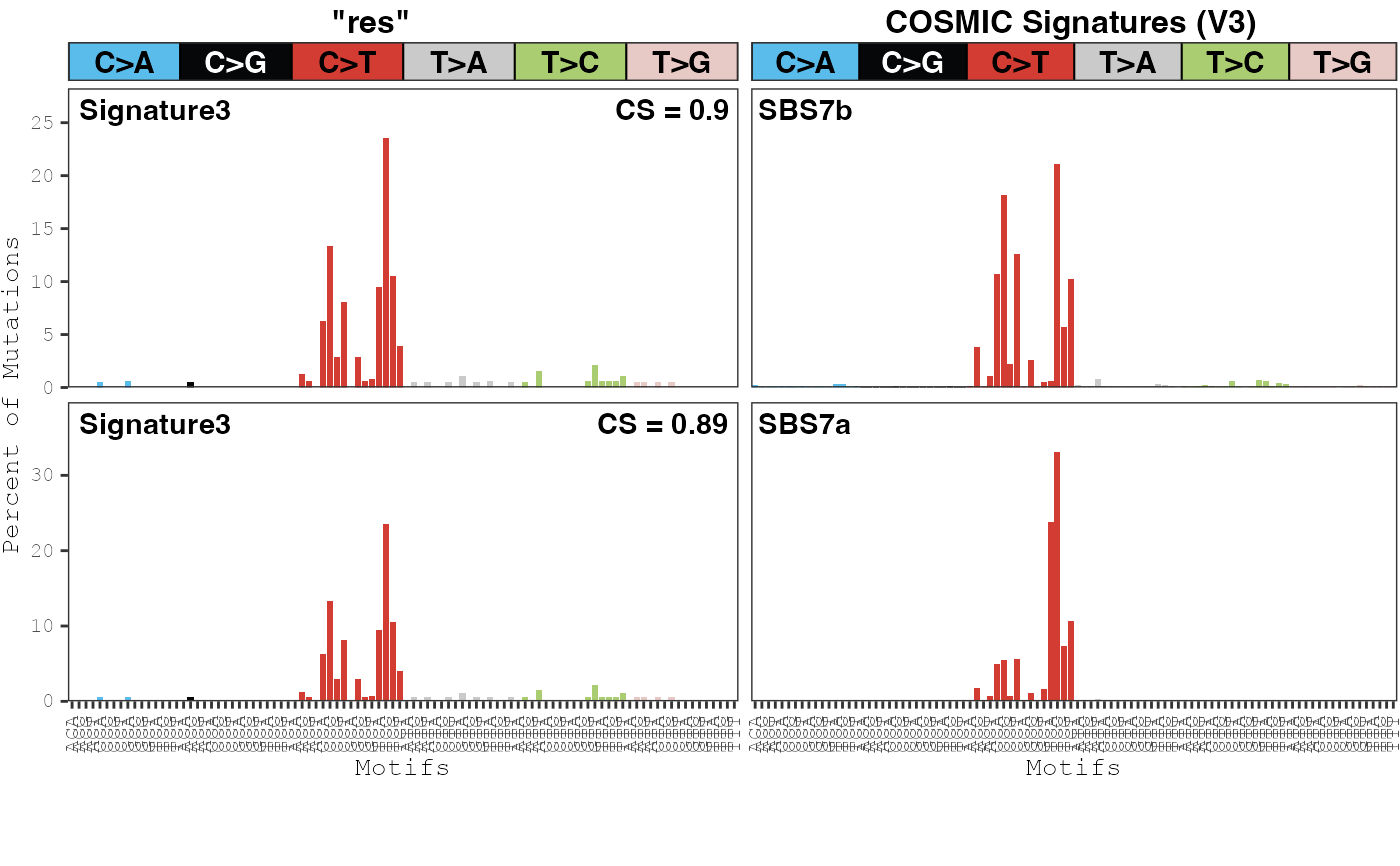 #> cosine x_sig_index y_sig_index x_sig_name y_sig_name
#> 2 0.9028629 3 8 Signature3 SBS7b
#> 1 0.8877524 3 7 Signature3 SBS7a
#> cosine x_sig_index y_sig_index x_sig_name y_sig_name
#> 2 0.9028629 3 8 Signature3 SBS7b
#> 1 0.8877524 3 7 Signature3 SBS7a