A wrapper function which visualizes outputs from the
runEmptyDrops function stored in the colData slot of the
SingleCellExperiment object.
plotEmptyDropsResults(
inSCE,
sample = NULL,
combinePlot = "all",
fdrCutoff = 0.01,
defaultTheme = TRUE,
dotSize = 0.5,
titleSize = 18,
axisLabelSize = 18,
axisSize = 15,
legendSize = 15,
legendTitleSize = 16,
relHeights = 1,
relWidths = 1,
samplePerColumn = TRUE,
sampleRelHeights = 1,
sampleRelWidths = 1
)Arguments
- inSCE
Input SingleCellExperiment object with saved dimension reduction components or a variable with saved results from
runEmptyDrops. Required.- sample
Character vector or colData variable name. Indicates which sample each cell belongs to. Default
NULL.- combinePlot
Must be either
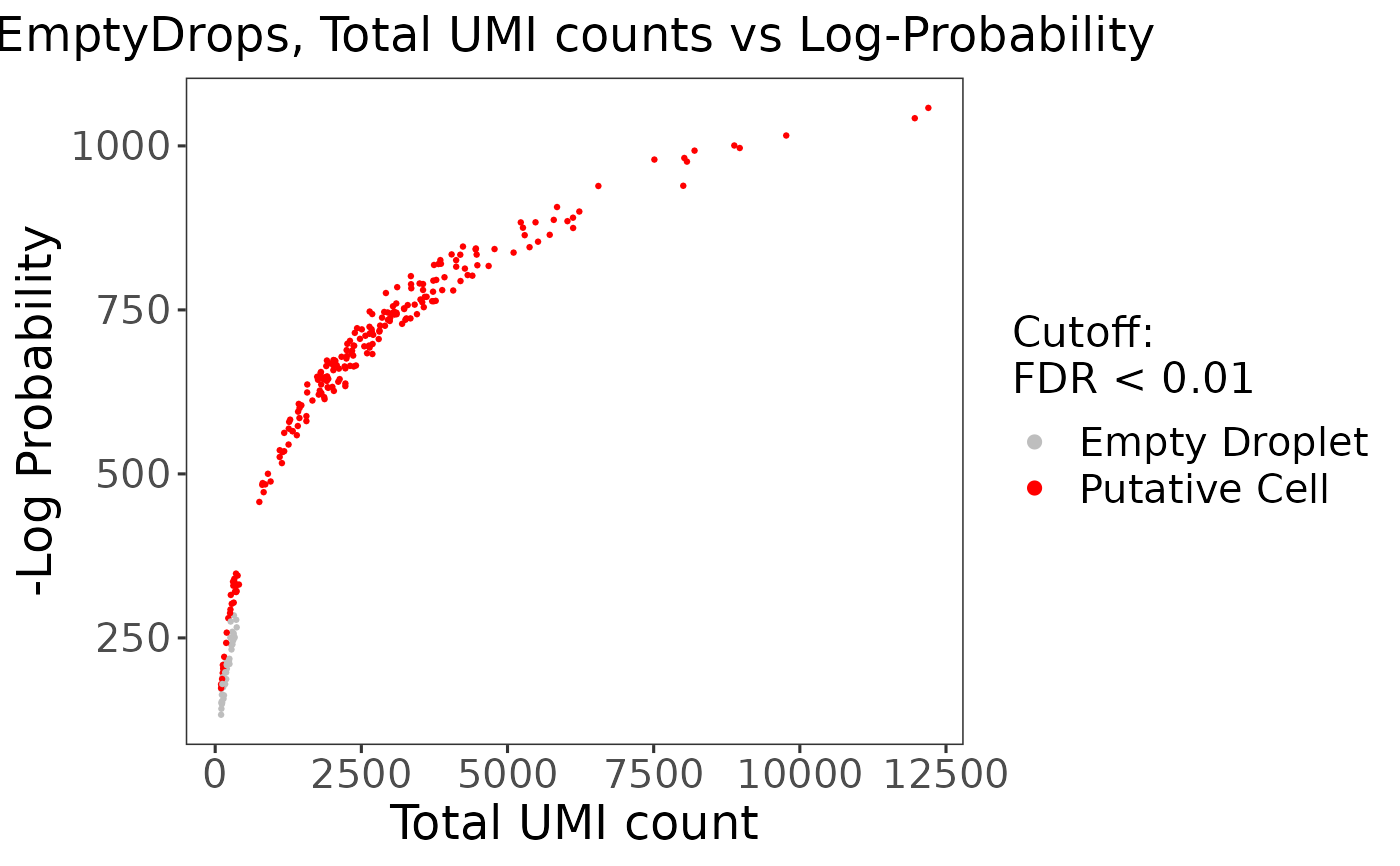
"all","sample", or object,"none"."all"will combine all plots into a single .ggplot while"sample"will output a list of plots separated by sample. Default"all".- fdrCutoff
Numeric. Thresholds barcodes based on the FDR values from
runEmptyDropsas "Empty Droplet" or "Putative Cell". Default0.01.- defaultTheme
Removes grid in plot and sets axis title size to
10whenTRUE. DefaultTRUE.- dotSize
Size of dots. Default
0.5.- titleSize
Size of title of plot. Default
18.- axisLabelSize
Size of x/y-axis labels. Default
18.- axisSize
Size of x/y-axis ticks. Default
15.- legendSize
size of legend. Default
15.- legendTitleSize
size of legend title. Default
16.- relHeights
Relative heights of plots when combine is set. Default
1.- relWidths
Relative widths of plots when combine is set. Default
1.- samplePerColumn
If
TRUE, when there are multiple samples and combining by"all", the output .ggplot will have plots from each sample on a single column. DefaultTRUE.- sampleRelHeights
If there are multiple samples and combining by
"all", the relative heights for each plot. Default1.- sampleRelWidths
If there are multiple samples and combining by
"all", the relative widths for each plot. Default1.
Value
list of .ggplot objects
See also
Examples
data(scExample, package = "singleCellTK")
sce <- runEmptyDrops(inSCE = sce)
#> Sat Mar 18 10:29:11 2023 ... Running 'emptyDrops'
plotEmptyDropsResults(inSCE = sce)
#> $scatterEmptyDrops
 #>
#>